The first thing in common between Operational Research (OR) and Biostatistics, is that both terms are often misunderstood. OR, or the Science of Better, as it is commonly known, has lots to do with optimization but there is much more to it…Some might be surprised to read that many of the tools of this discipline can be -and are actually- applied to problems in Biostatistics.
It all starts with Florence Nightingale volunteering as a nurse in the Crimean war in 1853…Her pioneering use of statistics for evidence-based medicine, and her operational research vision to reform nursing, led to a decline in the many preventable deaths that occurred throughout the nineteenth century in English military and civilian hospitals.
Since then, integrated approaches are commonplace: disease forecasting models, neural networks in Epidemiology, discrete-event simulation in clinical trials,…
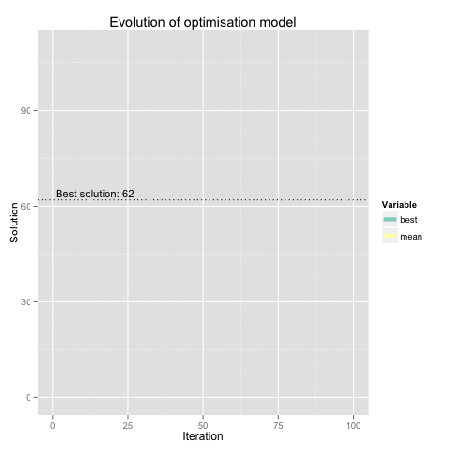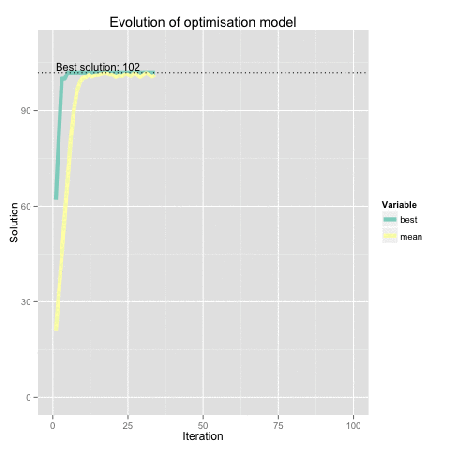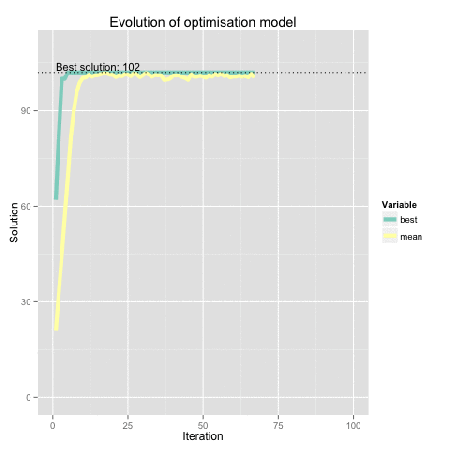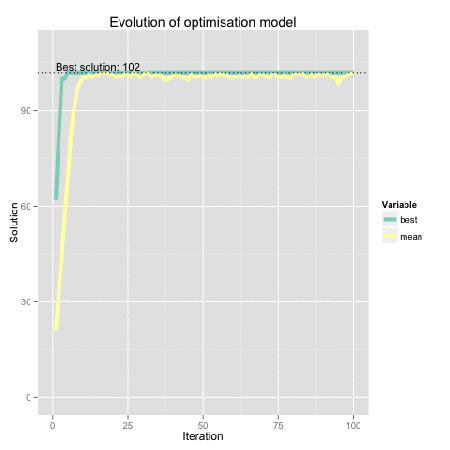
Figure: Genetic algorithm representation using R. Find here the code for the original dynamical plot by FishyOperations.
There is also an increasing interest in computational biology in OR. Examples of application of these techniques vary from linear programming based flux balance analysis models for cell metabolism studies, to sparse network estimation in the area of Genetics.
In R, there are many packages with which to apply these techniques, from genalg for Genetic algorithms (used to recreate figure above) or games on Games theory, to more specific packages in Bioconductor like CellNOptR, survcomp or OLIN. Also, a general Task View on Optimization can be found here.
Finally, a quick mention to the YoungOR 18 Conference that will be held in Exeter (UK), 9-11 April 2013. It will be covering topics of common interest for biostatisticians with streams on Health, Sustainability, etc. Plenty of inspiration for future posts!
Have you ever used any of these techniques?Any particular tips that you want to share?Tell us about it!