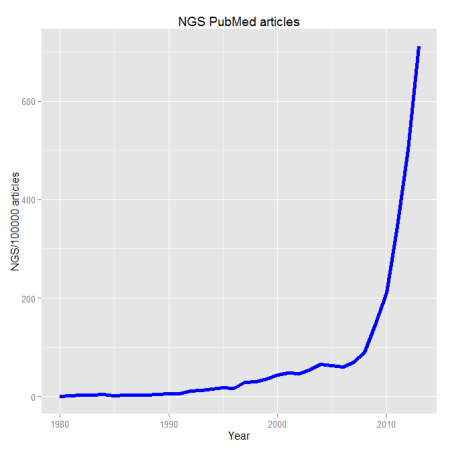
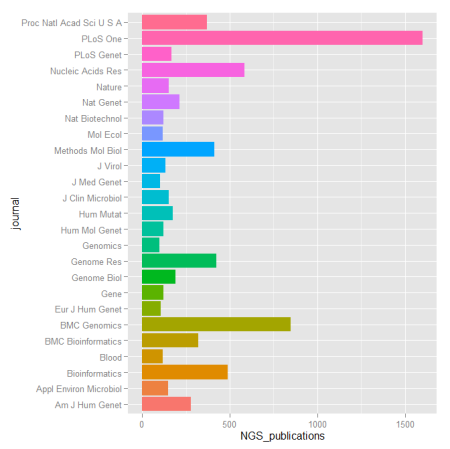
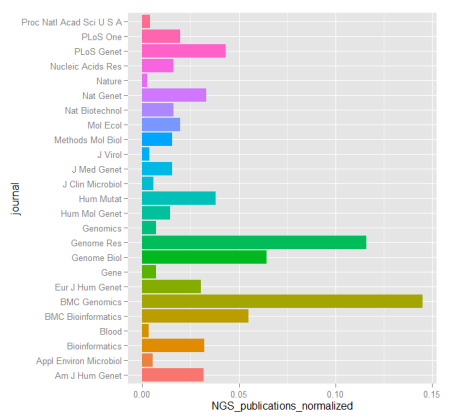
When dealing with genomic data, retrieving information from databases is a required endeavor. Most of these repositories of genomic and proteomic data have very useful and friendly interfaces or browsers perfectly suitable for particular searches. But if you are working with high dimensional data e.g. NGS data, more efficient tools to mine those databases are required. R offers several packages that make this task straightforward.
NCBI2R is an R package to annotate lists of SNPs, genes and microsatellites. It obtains information from NCBI databases. This package provides quite useful functions for getting a quick glance at a given set of SNPs or genes, retrieving SNPs in a gene or getting genes belonging to a KEGG pathway (among some other features) as shown in the code below.
library(NCBI2R)
GetIDs("MAPT[sym]")
GetGeneInfo(4137)
mysnps <- GetSNPsInGenes(4137)
GetSNPInfo(mysnps)
myset <- GetIDs("KEGG pathway:Alzheimer´s disease")
biomaRt package, part of Bioconductor project, is an R interface to the well-known BioMart data management system that provides access to several data sources including Ensemble, HGNC, InterPro, Reactome and HapMap.
Most of analysis can be performed with just two functions: useMart() to define the dataset, and getBM() to perform the query. It performs onlique queries baed on attributes – values we are interested in retrieving-, filters – restrictions on the query- and a given set of values of the filter. Once we define the dataset we are interested in, we can check all the filters and attributes available. Let´s say we want to look for those genes associated with an OMIM phenotype.
source( "http://www.bioconductor.org/biocLite.R")
biocLite("biomaRt")
library(biomaRt)
listMarts()
mymart <- useMart("ensembl",dataset = "hsapiens_gene_ensembl")
listFilters(mymart)
listAttributes(mymart)
getBM(attributes = c("entrezgene", "hgnc_symbol", "mim_morbid_description"), filters ="entrezgene", values = myset, mart = mymart)
Org.Hs.eg.db , which is also part of Bioconductor, is an organism specific package that provides annotation for the human genome. It is based on mapping using Entrez Gene identifiers. In a very similar way to biomaRt, it allows to query the databases by means of the select() function specifying cols – kind of data that can be returned-, keytypes -which of the columns can be used as keys- and a given key.
source("http://bioconductor.org/biocLite.R")
biocLite("org.Hs.eg.db")
library(org.Hs.eg.db)
keytypes(org.Hs.eg.db)
cols(org.Hs.eg.db)
select(org.Hs.eg.db, keys= "MAPT", cols = c("SYMBOL", "CHR", "CHRLOC", "UNIPROT"), keytype = "SYMBOL")
These three packages offer in many cases a good alternative to other well-known data mining systems such as UCSC Table Browser data retrieval tool. Besides, commands from these packages are quite simple, so even those not that familiar with R language can take good benefit from them.
Just a quick tip to finish – I have just found out about fread() function (data.table package) for reading large files. It really makes a difference!